Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Pippione, A.C., Carnovale, I.M., Bonanni, D., Sini, M., Goyal, P., Marini, E., Pors, K., Adinolfi, S., Zonari, D., Festuccia, C., Wahlgren, W.Y., Friemann, R., Bagnati, R., Boschi, D., Oliaro-Bosso, S., Lolli, M.L.(2018) Eur J Med Chem 150: 930-945
- PubMed: 29602039
- DOI: https://doi.org/10.1016/j.ejmech.2018.03.040
- Primary Citation of Related Structures:
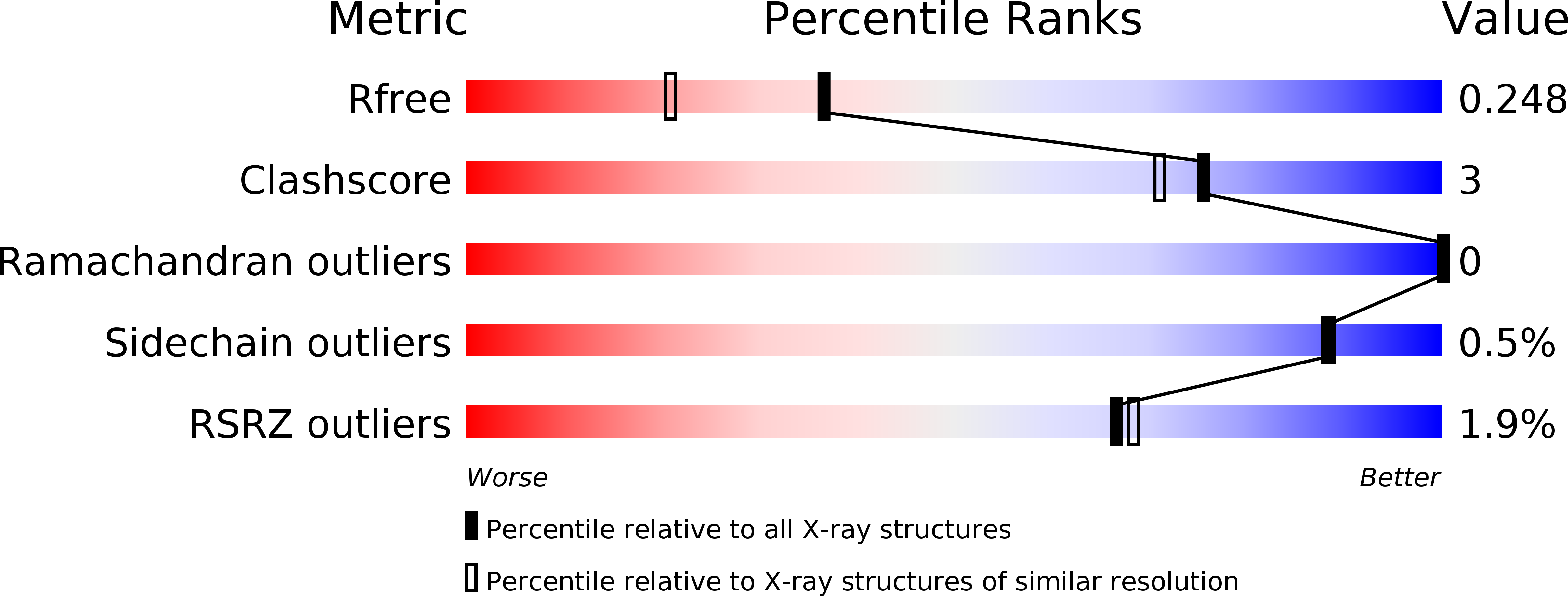
6F2U, 6F78 - PubMed Abstract:
The aldo-keto reductase 1C3 (AKR1C3) isoform plays a vital role in the biosynthesis of androgens and is considered an attractive target in prostate cancer (PCa). No AKR1C3-targeted agent has to date been approved for clinical use. Flufenamic acid and indomethacine are non-steroidal anti-inflammatory drugs known to inhibit AKR1C3 in a non-selective manner as COX off-target effects are also observed. Recently, we employed a scaffold hopping approach to design a new class of potent and selective AKR1C3 inhibitors based on a N-substituted hydroxylated triazole pharmacophore. Following a similar strategy, we designed a new series focused around an acidic hydroxybenzoisoxazole moiety, which was rationalised to mimic the benzoic acid role in the flufenamic scaffold. Through iterative rounds of drug design, synthesis and biological evaluation, several compounds were discovered to target AKR1C3 in a selective manner. The most promising compound of series (6) was found to be highly selective (up to 450-fold) for AKR1C3 over the 1C2 isoform with minimal COX1 and COX2 off-target effects. Other inhibitors were obtained modulating the best example of hydroxylated triazoles we previously presented. In cell-based assays, the most promising compounds of both series reduced the cell proliferation, prostate specific antigen (PSA) and testosterone production in AKR1C3-expressing 22RV1 prostate cancer cells and showed synergistic effect when assayed in combination with abiraterone and enzalutamide. Structure determination of AKR1C3 co-crystallized with one representative compound from each of the two series clearly identified both compounds in the androstenedione binding site, hence supporting the biochemical data.
Organizational Affiliation:
Department of Science and Drug Technology, University of Torino, via Pietro Giuria 9, 10125, Torino, Italy.